After the job is done, you will see a summary webpage similar as in the following figure.
The result files would be found in the Status section.
In addition, you can see information about the workflow run from the other sections, such as input files used, the job status, running time and its computation graph, etc.
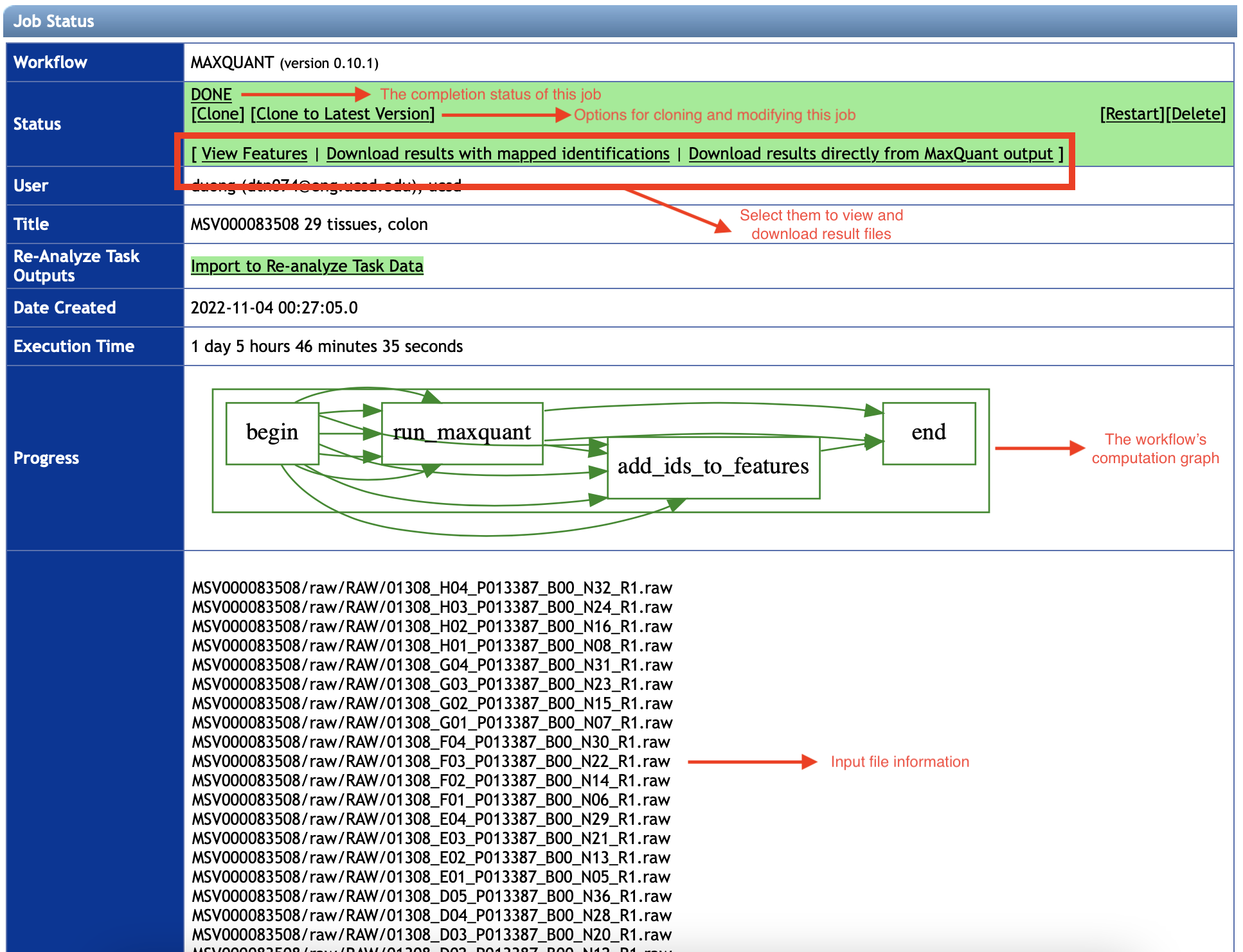
3.1 View Features and Download MaxQuant Result Files
Once you select View Features button in the Status section, the workflow will provide a list of features detected and mapped to Andromeda's peptide IDs by MaxQuant software.
This result is basically from the text file evidence.txt that you could find in the directory /combined/txt if running MaxQuant locally.
If you are interested in a complete set of MaxQuant result files including allPeptides.txt, evidence.txt, peptides.txt, proteinGroups.txt, etc.,
please use the button Download results directly from MaxQuant output, then click the Download in the top-right corner.
The description of these files can be found in the MaxQuant tutorial.
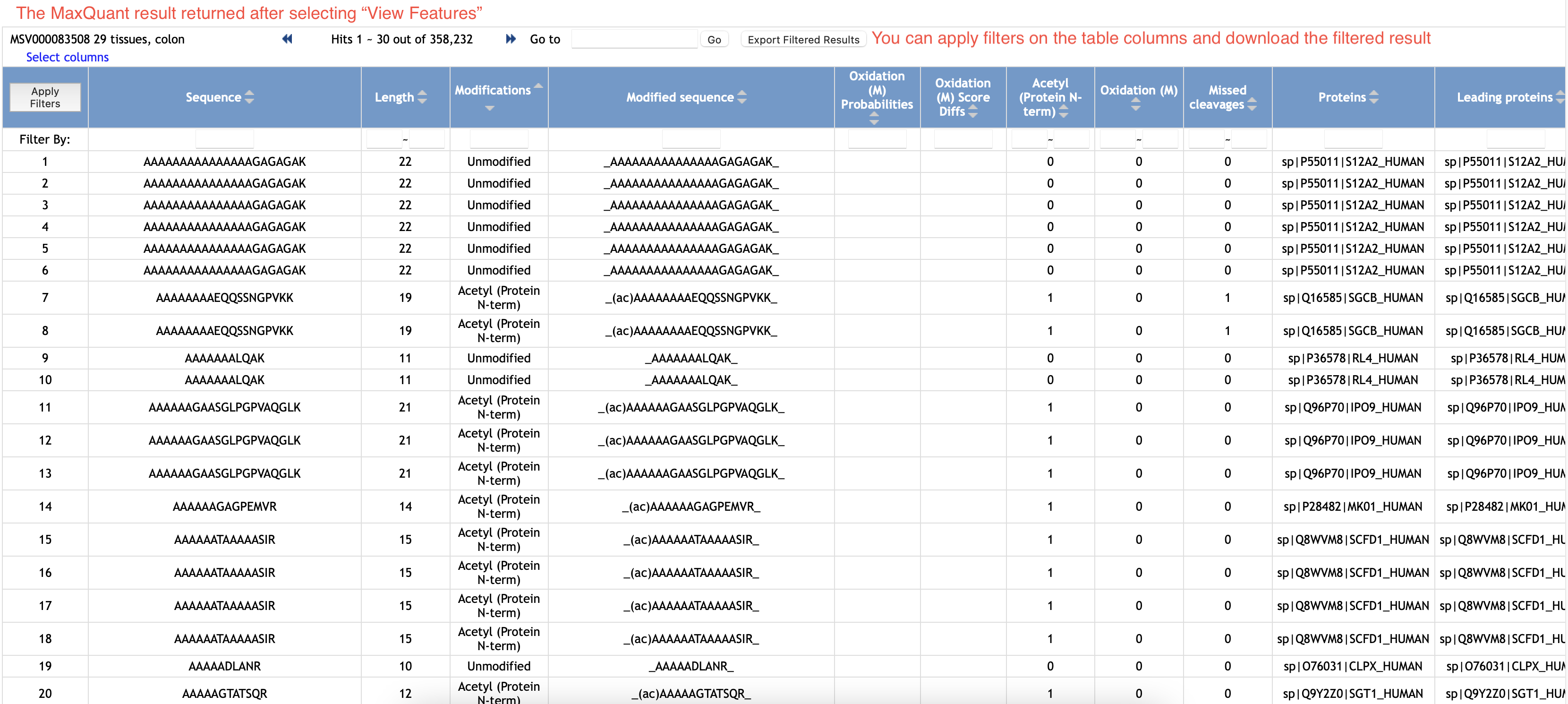
3.2 Download Result Files by Our Advanced Mapping
If you provided peptide IDs in the section Advanced Identification Mapping Parameters before starting your workflow,
you can find the results with the mapped IDs by the option Download results with mapped identifications in the Status section.
The information of mapped features, peptides and proteins is stored in .tsv files in the downloaded folder mapped_features.
The file evidence.tsv would be the most important file among them. This probably contains all information needed for your further analyses.
Our mapping also outputs evidence.tsv and proteins.tsv as the same formats with MaxQuant's evidence.txt and proteinGroups.txt, respectively.
Thus, you can easily read our mapping and MaxQuant's result files in the same way.