FBMN with MS-DIAL
1. Introduction¶
The main documentation for Feature-Based Molecular Networking can be accessed here. See our article.
Below we are describing how to use MS-DIAL with the FBMN workflow on GNPS.
2. Mass spectrometry processing with MS-DIAL¶
Download the latest version of MS-DIAL software at http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL.
Citations and development¶
Recommended Citations
This work builds on the efforts and tools from our many colleagues, please cite their work:
Nothias, L.-F., Petras, D., Schmid, R. et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 17, 905–908 (2020).
Wang, M. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016).
Tsugawa, H., et al A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 38, 1159–1163 (2020).
Tsugawa, H., Cajka, T., Kind, T., Ma, Y., Higgins, B., Ikeda, K., Kanazawa, M., VanderGheynst, J., Fiehn, O. & Arita, M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523-526 (2015).
Lai, Z., Tsugawa, H., Wohlgemuth, G., Mehta, S., Mueller, M., Zheng, Y., Ogiwara, A., Meissen, J., Showalter, M., Takeuchi, K., Kind, T., Beal, P., Arita, M. & Fiehn, O. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 15, 53-56 (2018).
Mass Spectrometry Data Processing with MS-DIAL¶
In MS-DIAL, a sequence of steps is performed to process the mass spectrometry data. Here we will present key steps required to process LC-MS/MS data acquired in non-targeted mode (data dependent acquisition). MS- DIAL can process LC-MS/MS data performed in non-targeted mode, but also in MSE and now with Ion Mobility Spectrometry. For convenience we also provide a batch file (XML format) that can be imported directly in MS-DIAL.
IMPORTANT: MS-DIAL parameters will vary depending on the mass spectrometer, the acquisition parameters, and the samples studied. In this page we present a basic guideline for using MS-DIAL with the FBMN on GNPS.
Documentation¶
Please consult these resources for more details on MS-DIAL processing:
- See the MS-DIAL documentation on LC-MS/MS data processing
- See the MS-DIAL documentation for MSE data processing
- See the MS-DIAL documentation on Ion Mobility Spectrometry data processing
Video Presentation and Tutorials¶
- The presentation/tutorial about MS-DIAL from the Leibniz Institute of Plant Biochemistry.
- This video from the WCMC about MS-DIAL
- The video tutorial about MS-DIAL processing for FBMN on GNPS.
Convert your LC-MS/MS Data to Open Format¶
MS-DIAL accepts different input formats. Note that we recommend first to convert your files to mzML format before doing MS-DIAL processing. See the documentation here.
MS-DIAL releases later than ver 4.60 supports direct import of SCIEX (.wiff, .wiff2), Thermo (.raw). Agilent (.d), and Waters (.raw), so conversion is not required. See the release note
Processing Steps for non-targeted LC-MS/MS¶
Below is a walk-through of all the steps
1. Make a New Project¶
Go to Menu: File / New Project
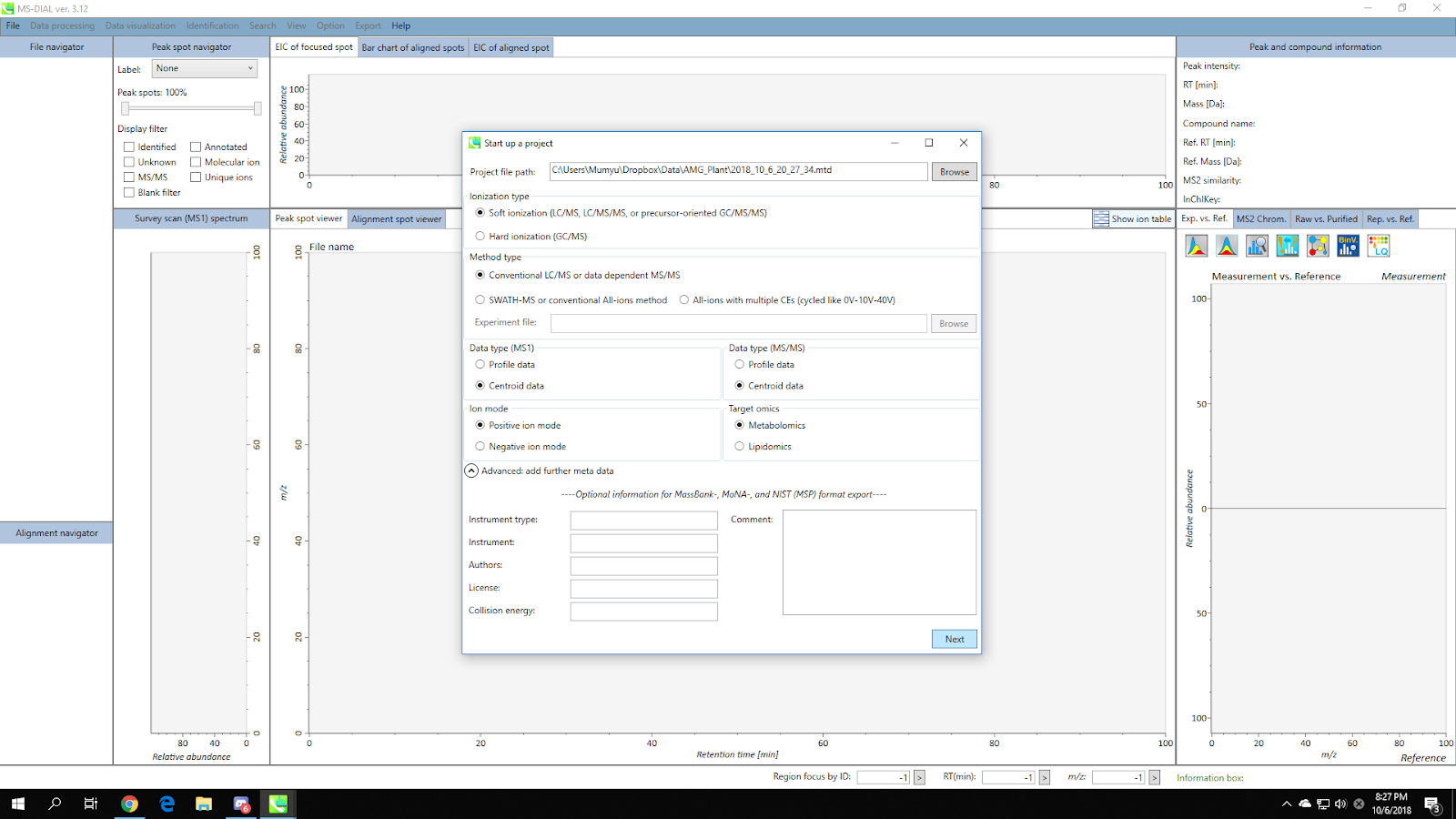
Select appropriate parameters (ionization type / method type / data type / ion mode / target omics) for your dataset. Click “Next”.
FBMN with Waters MSe data
If you are processing with Waters MSe data, set MS method type to 'SWATH-MS or conventional All-ions method' and use this experiment file.
2. Import Files¶
Click “browse” and open the dropdown menu for file types, and select “mzML file(*.mzml)”. The other available options are .abf or .cdf files.
[Note] Data files should be placed at the same folder with the project file.
Select files and click “Open”. You will see the list of selected files. Click “Next”.
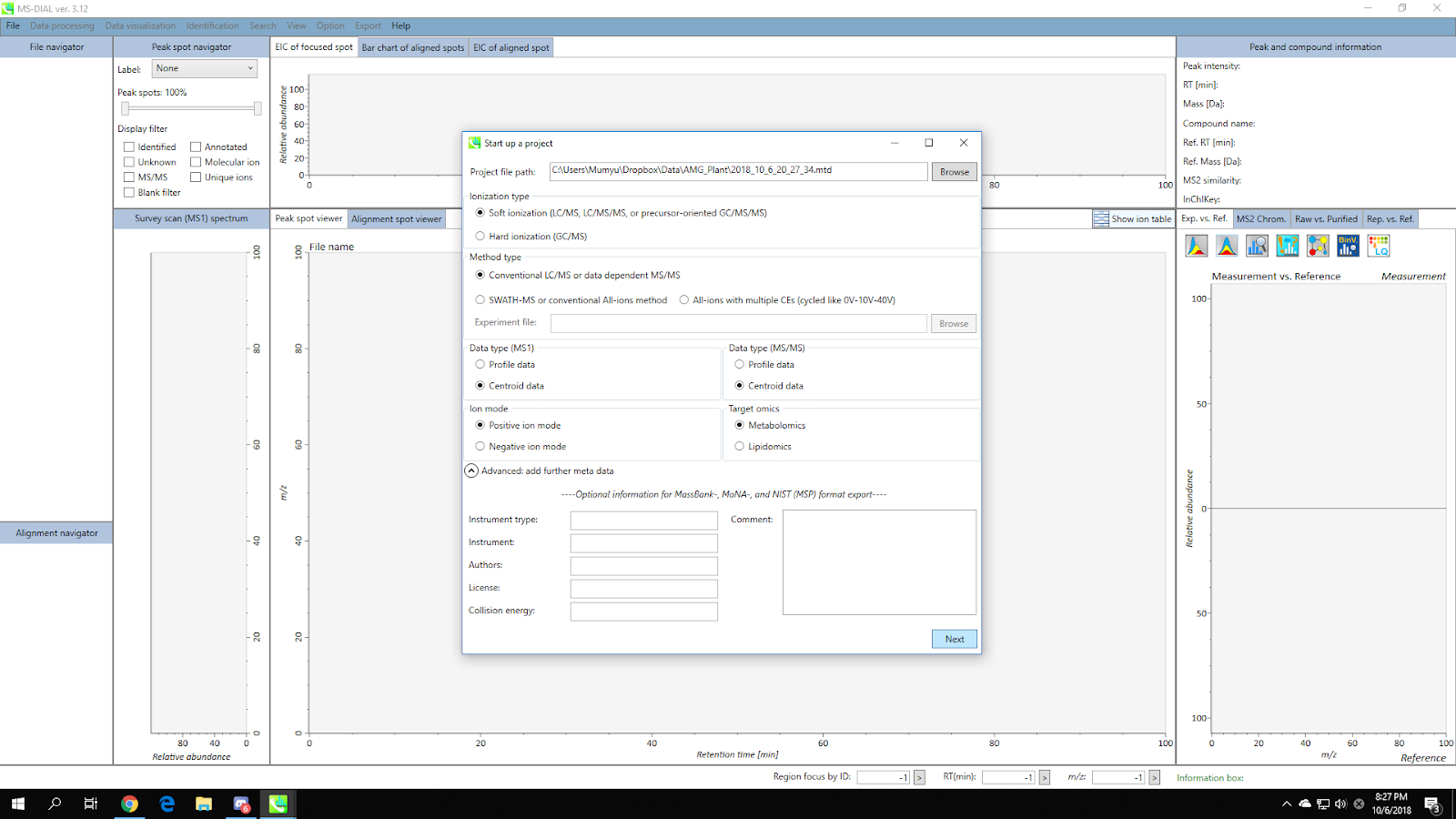
3. Set Parameters - Data Collection¶
Set the MS1/MS2 tolerance and data collection parameters.
If you are dealing with a large dataset, you can reduce the running time by setting multithreading option in the “Advanced” menu.
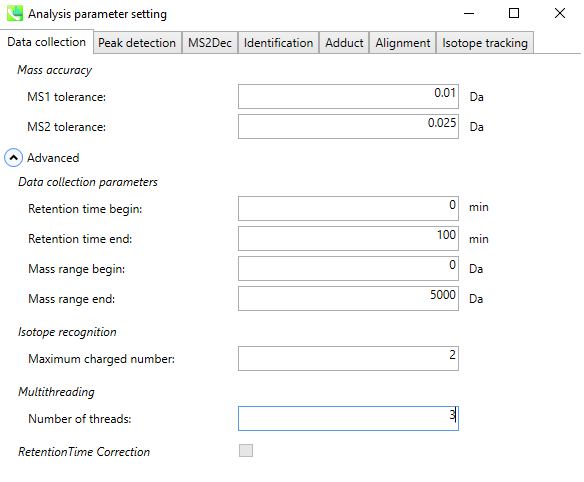

4. Set Parameters - Peak Detection¶
Set the MS1 peak detection parameters. You should set a peak height threshold that is adapted to the mass spectrometer.
As a rule of thumb, the value should at least correspond to the minimum value set for the triggering of the MS2 scan event.
If you want to remove some specific ions (e.g. known contaminants), you can make an exclusion list here.
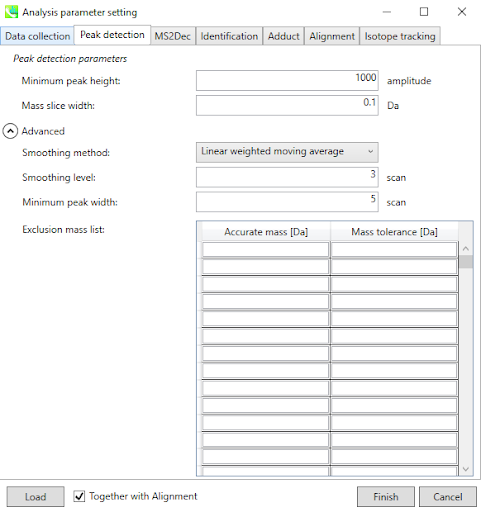
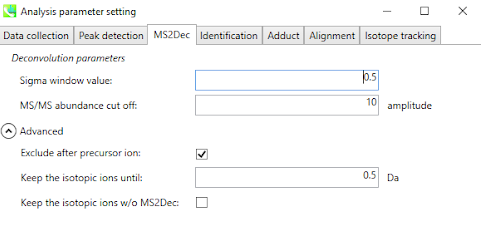
5. Set Parameters - MS2Deconvolution¶
You should set an appropriate MS/MS abundance cutoff. Make sure to set an intensity threshold representative of the noise level in MS2 spectra.
This is typically lower than for MS1. If you have any doubt, set it to 0.

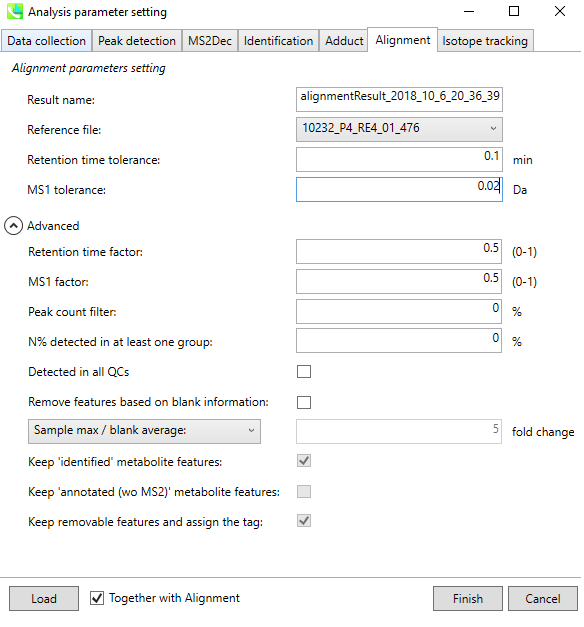
6. Set Parameters - Alignment¶
Set appropriate Retention time / MS1 tolerance for alignment.
If you have any quality control (QC) sample data, it is a nice candidate to be the reference file.
Click “Finish”, then MS-DIAL will perform all the feature detection and alignment process.
Processing Steps for MSE data¶
-
See the MS-DIAL documentation for MSE data processing, and export the files as indicated below. If you encounter any issue, please contact the GNPS team on the forum or GitHub.
-
Here is an example MSe-based MS-DIAL networking job. MSe .Raw and converted ABF files can be downloaded in here.
Ion Mobility Spectrometry data (Experimental)¶
- See the MS-DIAL documentation on Ion Mobility Spectrometry data processing, and export the files as indicated below. If you encounter any issue, please contact the GNPS team on the forum or GitHub.
Export the files for GNPS FBMN¶
To run FBMN after MS-DIAL processing, two files have to be exported:
-
The feature quantification table (TXT file) containing compound intensity and annotation.
-
The MS/MS spectral summary (MGF file) containing the list of representative MS/MS spectra (the most intense MS/MS spectrum).
Go to Menu: Export / Alignment result
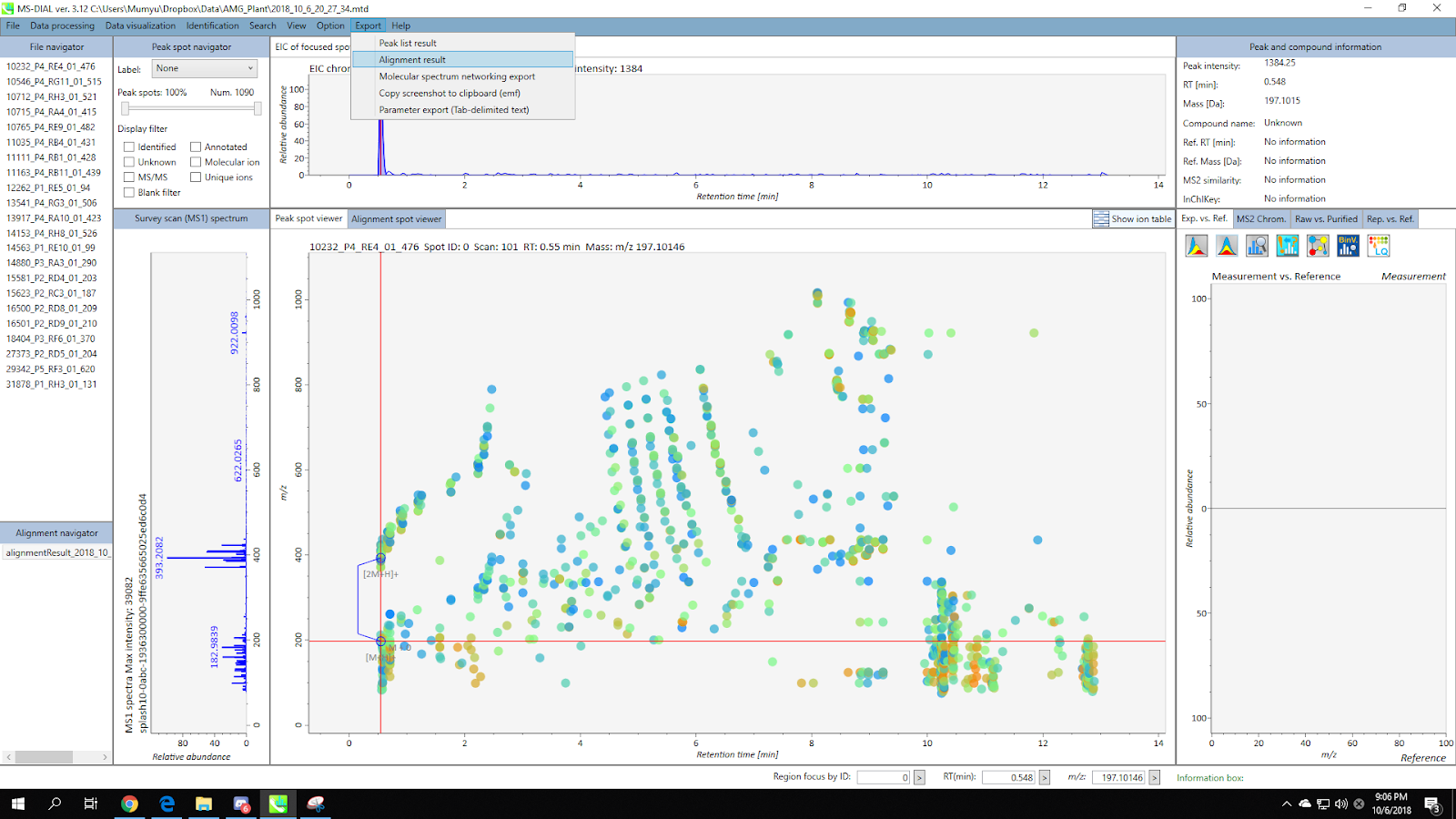
Make sure the “GNPS export” box is checked, and set the spectra type to “centroid”.
Set the directory and file name, then click “Export”. As a result, you will acquire a MS/MS spectral summary and the feature quantification table.
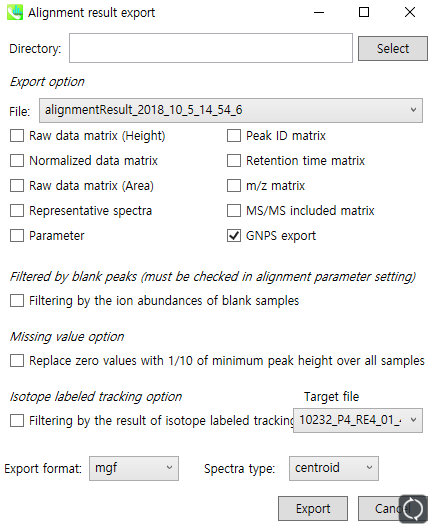
[Note] The GNPS export option of MS-DIAL will create a feature table based on MS1 peak height. If you want a feature table based on peak area, check the “Raw data matrix (Area)”. The resulting file has a different structure from TXT file given by GNPS export, so you should reformat the area matrix file.
See example of files generated by MS-DIAL and used for the FBMN by the export workflow here.
3. Running the FBMN with MS-DIAL¶
After the processing with MS-DIAL, the output files can be used to run the Feature-Based Molecular Networking workflow on GNPS either using the [Superquick FBMN start page] (http://dorresteinappshub.ucsd.edu:5050/featurebasednetworking) or the standard interface of the FBMN workflow (you need to be logged in GNPS first).
See the main documentation for more details about the FBMN workflow on GNPS.
Basically, you will need to upload the files produced by MS-DIAL:
- The feature quantification table (.TXT file)
- The MS/MS spetral summary file (.MGF file)
- (Optional) The metadata table - requirements are described here.
IMPORTANT WHEN USING A METADATA TABLE WITH MS-DIAL: "filename" in the metadata files for FBMN with MS-DIAL should not contain file extensions (ex: "sample1.mzML" must be "sample1").
Here is an example MS-DIAL networking job from a subset of the American Gut Project.
Tutorials¶
See our MS-DIAL tutorial on using Feature Based Molecular Networking for the American Gut Project sample.
Join the GNPS Community !¶
- For feature request, or to report bugs, please open an "Issue" on the CCMS-UCSD/GNPS_Workflows GitHub repository.
- To contribute to the GNPS documentation, please use GitHub by forking the CCMS-UCSD/GNPSDocumentation repository, and make a "Pull Request" with the changes.
=======
Page Contributions¶