Merge Network Polarity
Here we merge polarity between positive and negative networks.
Workflow¶
The workflow is available on GNPS here
The merge polarity workflow can be performed on both classical and feature based molecular networking (FBMN) jobs.
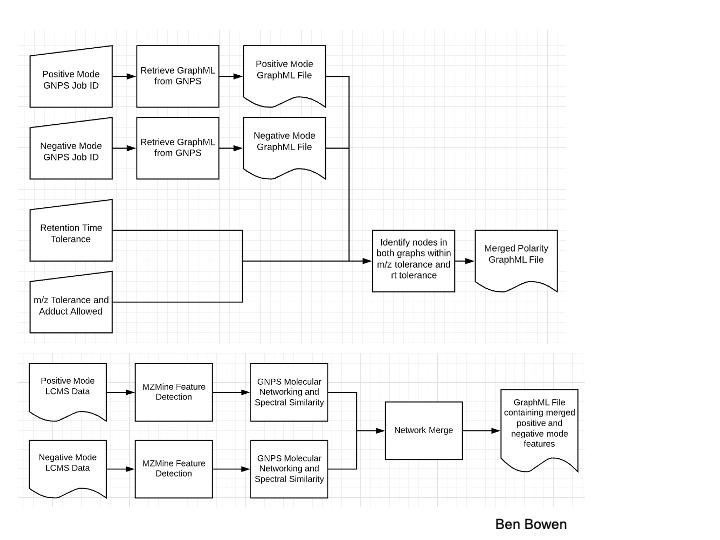
A GNPS job is run on positive mode data and another job is run on negative mode data; the input is then the GNPS ID of both jobs. These jobs can be either classical or feature based molecular networking (FBMN) jobs.
A retention time tolerance for aligning masses between the two jobs must be specified. Note that this time should be given in minutes for FBMN jobs and in seconds for classical molecular networking jobs. A ppm tolerance must also be specified for the merge.
Interpreting Results¶
A merged graphml file is created along with summary statistics and evaluations. To provide an assessment of accuracy of the edges inserted, new edges where both nodes in the respective positive and negative networks are identified with a structure (an annotation), a tanimoto score is calculated between the structures. Tanimoto scores range from 0-1 when the nodes from positive and negative jobs are annotated by library matching, where 1 represents perfect concordance of structure while 0 represents no concordance. If no structures are annotated, the tanimoto score is given as -1. We assume a tanimoto score of greater than 0.9 indicates concordance of structure.
Citation¶
Source code is available on Github
Page Contributors¶